May 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Waspergillamide A
Aspergillus sp. (CMBW031) is a fungus isolated from the mud dauber wasp found in Brisbane, Australia. It was found that it was an undocumented strain related to Apergillus westerdijkia NRRL 3174. In order to explore its secondary metabolite behavior, the fungus was subjected to cultivations under 38 different conditions. HPLC-DAD-ESIMS analysis of the solvent extracts revealed a diverse range of secondary metabolites. Quezada et al [1] did investigations of CMB-W031 in rice grain cultivations. After reverse-phase fractionation a new nitro depsi-tetrapeptide diketopiperazine, waspergillamide A (1), was isolated along with the known metabolites. The structure elucidation of it is described in [1].
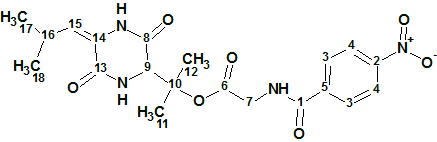
1
HR-ESI(+)MS measurements on 1 showed a sodium adduct ([M + Na]+ with a molecular formula of C20H24N4O7 (DBE=11). Initial inspection of the 1D NMR data indicated the presence of a tri-substituted olefin, four amide/ester carbonyls, and a p-di-substituted benzene. A UV−Vis spectrum (λmax 275 nm, log ε 3.73), and investigated NMR data, supported the assignment of Δ2,3- Leu, 3-OH-Val, Gly, and p-nitrobenzene residues.
To verify the proposed structure assignment, a sample of 1 was hydrolyzed overnight in 6 M HCl at 100 Co to produce a hydrolysate, from which HPLC-DAD-ESIMS analysis detected the predicted ester hydrolysis products.
Authors [1] omitted mentioning the considerations that led them to conclude an NO2 group is present in the molecule. It is known[2] that losses of 46 (NO2) and 30 (NO) amu in the MS spectrum, and two strong IR bands close to 1530 and 1350 cm-1, are the features of the presence of an NO2 group. Unfortunately neither MS nor IR spectra are available from [1]. Nevertheless, we assume that the authors in [1] confirmed the presence of the nitro group in the molecule.
The NMR data used in [1] for the structure elucidation are presented in Table 1.
Table 1. Waspergillamide A. Spectroscopic NMR data
| Label | δC | δC calc* | XHn | δH | M | COSY | H to C HMBC |
| C 1 | 168.9 | 166.39 | C | ||||
| C 2 | 151.7 | 149.63 | C | ||||
| C 3 | 130.3 | 129.03 | CH | 8.07 | d | 8.33 | C 2, C 1 |
| C 4 | 125 | 123.92 | CH | 8.33 | d | 8.07 | C 5, C 2 |
| C 5 | 140.8 | 139.95 | C | ||||
| C 6 | 170 | 169.04 | C | ||||
| C 7 | 43.6 | 42.39 | CH2 | 4.02 | s | C 1, C 6 |
|
| C 8 | 165.6 | 163.63 | C | ||||
| C 9 | 63.7 | 60.64 | CH | 4.34 | s | C 10, C 13, C 8 |
|
| C 10 | 86.4 | 85.05 | C | ||||
| C 11 | 23.8 | 24.56 | CH3 | 1.63 | s | C 12, C 9, C 10 |
|
| C 12 | 23 | 29.03 | CH3 | 1.54 | s | C 11, C 9, C 10 |
|
| C 13 | 163.9 | 163.18 | C | ||||
| C 14 | 126.9 | 123.46 | C | ||||
| C 15 | 128.2 | 128.3 | CH | 5.85 | d | ||
| C 16 | 26.4 | 27.44 | CH | 2.75 | u | 1.05, 1.07 |
C 17, C 13 |
| C 17 | 22.7 | 23.02 | CH3 | 1.07 | d | 2.75 | |
| C 18 | 22.9 | 23.02 | CH3 | 1.05 | d | 2.75 | C 14 |
* 13C chemical shift calculations were carried out using a HOSE code based approach [2]
The data collected in Table 1 were input into ACD/Structure Elucidator and a Molecular Connectivity Diagram (MCD) was produced by the program. The MCD edited by the user is shown in Figure 1.
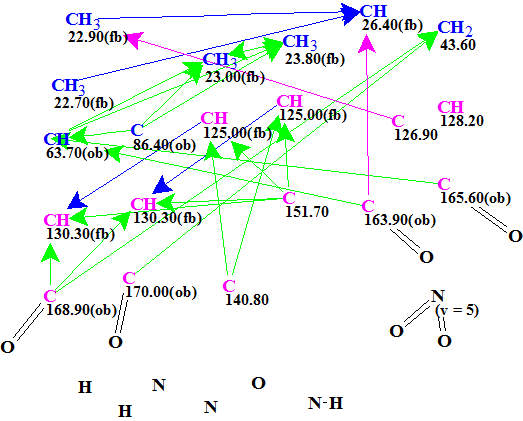
Figure 1. Edited molecular connectivity diagram.
MCD overview. Even though the molecule is of a modest size (C20H24N4O7), it is still a challenge for CASE because the molecular formula implies DBE=11, and it contains four nitrogen atoms that show variable valences. Strictly speaking, if we tried to solve the problem in an “ab intio” mode we would be forced to perform structure generation with the option “Enumerate possible valences for N, S and P atoms”, which would have investigated all the valence options combinations for the 4 nitrogen atoms (5 per nitrogen). In this case the processing time necessary for searching of all possible valence combinations for all nitrogen atoms during the structure generation would take a long time. Therefore we assumed the presence of one NO2 group, which was drawn in MCD manually. To further confirm the possibility of an NO2 present, we performed 13C chemical shift search against the Fragment Library incorporated within the ACD/Structure Elucidator. As a result of subsequent Functional Group search in the Found Fragments it was revealed that 65 of 2341 Found Fragments contain NO2, which indirectly hints at the presence of an NO2.
At least one NH group was detected by the authors [1] and it was drawn manually. Unfortunately no HMBC pattern is shown in the supporting information of article [1], so visual evaluation of peak intensities in the spectrum is impossible. Because HMBC correlations 1.05/C14 and 2.75/C13 are of 4J length we supposed that their intensities allow one to assume the correlation lengths are 2-4 bonds (pink arrows in the MCD). Following the authors’ suggestion, four C=O groups were drawn in the MCD manually. Finally, signal multiplicities in 1H NMR shown in Table 1 (the column M) were assigned to corresponding atoms.
Strict Structure Generation accompanied with 13C chemical shift prediction and structure filtering was performed. The generated structures for which average deviations of 13C predicted chemical shifts from the experimental exceeded 5 ppm were rejected. The following results were obtained: 42680→468→320, tg = 1 m 53 s.
Furthermore, according to the CASE methodology, 13C chemical shift prediction was applied to all structures in the output file using additive rules, neural networks and fragmental (HOSE code-based) approaches [2], and average chemical shift deviations (dI, dN and dA) were calculated. All 320 structures were then ranked in increasing order of dA values. The six top-ranked structures are presented in Figure 2.
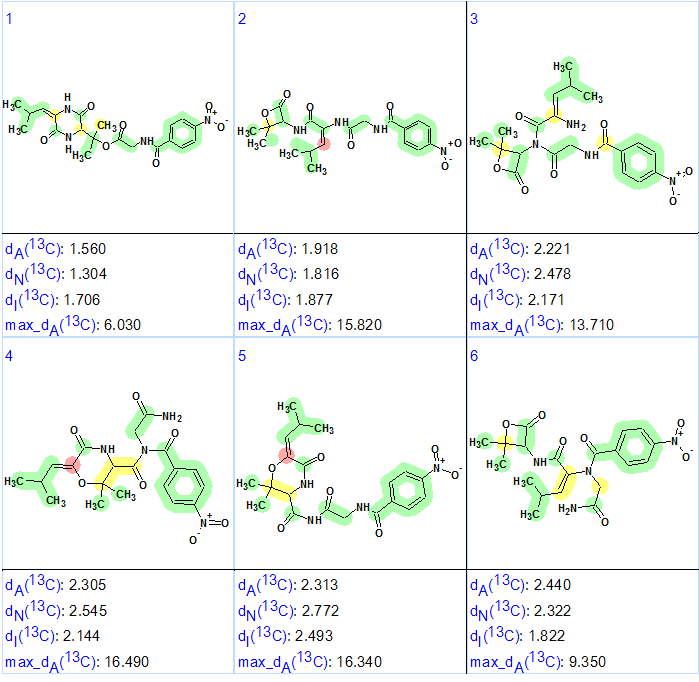
Figure 2. The six top-ranked structures of the output file. The color attributes of the 13C chemical shift prediction accuracy are: ±3 ppm—green, between 3 and 15 ppm—yellow, greater than 15 ppm—red.
Figure 2 shows that the top-ranked structure #1 is the same as that of of Waspergillamide A proposed in [1]. The deviations calculated, as well as the accuracy of the carbon atom chemical shift prediction, are marked with color and allow us to conclude that the solution obtained using ACD/Structure Elucidator Suite is correct. Though differences between deviations inherent for the first and second structures are small, the priority of the structure #1 is confirmed by the max_dA values. In addition all competing structures which contain a four-membered lactone would be easily rejected by the absence of a characteristic carbonyl band near 1780 cm -1 in the IR spectrum.
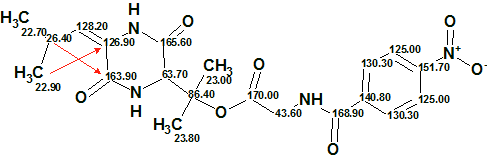
The elucidated structure with automatically assigned 13C chemical shifts is shown below. Red arrows show 4J HMBC correlations.
References
- M. Quezada, Z. Shang, P. Kalansuriya, A.A. Salim, E. Lacey, R.J. Capon. (2017). Waspergillamide A, a Nitro depsi-Tetrapeptide Diketopiperazine from an Australian Mud Dauber Wasp-Associated Aspergillus sp. (CMB-W031). J. Nat. Prod. 80 (4): 1192–1195.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


