January 1, 2018
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Zanthplanispine
Zanthoxylum planispinum. (Zhu-Ye- Jiao in Chinese) is a plant that can be found in the southeast and southwest regions of China. It is used in Chinese medicine as a painkiller/anti-inflammatory for toothache, stomachache and snake bites. It is also used for treating colds and expelling roundworms. This plant has been investigated previously and found to contain a lot of lignans, amongst which also were tetrahydrofuran and furofuran. Some of them shoed promising anti-inflammatory activity. In a search for novel anti-inflammatory bioactive drug leads from this plant, Su at al [1] isolated from its roots a novel tetrahydrofuran lignan named zanthplanispine (1), together with another fourteen known lignans.
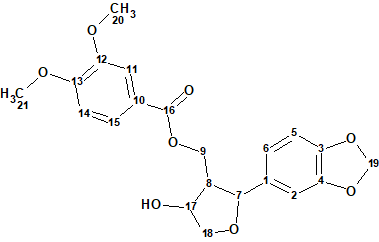
1
The structure of this compound was elucidated by spectroscopic methods, including 2D-NMR techniques. Compound 1 belongs to a rare and unusual type of tetrahydrofuran lignans, of which only two similar compounds have been previously reported.
Compound 1 was isolated as a colorless oil. The molecular formula was found to be C21H22O8 by its positive ion HR-ESI-MS at m/z 425.1218 ([M + Na]+, calc. 425.1207), which was in agreement with the 1H, 13C and DEPT NMR data. The IR spectrum showed absorption bands characteristic of the hydroxyl (3458 cm-1), carbonyl (1710 cm‑1), and aromatic (1590, 1506 cm-1) groups. The absorption bands in UV spectrum (221, 265 and 291 nm) also suggested the presence of the aromatic and unsaturated moieties.
The 1D and 2D NMR data (Table 1) utilized in [1] for structure elucidation were used for challenging ACD/Structure Elucidator.
Table 1. NMR spectroscopic data for compound
| Label | δC | δCcalc | CHn | δH | COSY | H to C HMBC |
| C 1 | 134.7 | 133.25 | C | |||
| C 2 | 106.9 | 104.42 | CH | 6.97 | C 4, C 6, C 7 | |
| C 3 | 148.8 | 147.61 | C | |||
| C 4 | 147.4 | 147.43 | C | |||
| C 5 | 108.2 | 106.73 | CH | 6.75 | 6.86 | C 1, C 4 |
| C 6 | 120.1 | 119.68 | CH | 6.86 | 6.75 | C 1, C 2, C 7 |
| C 7 | 83.3 | 84.41 | CH | 4.6 | 2.5 | C 1, C 2, C 6, C 8, C 9 |
| C 8 | 56.1 | 53.64 | CH | 2.5 | 4.40, 4.45, 4.60 | C 1, C 7, C 9, C 17 |
| C 9 | 63.2 | 61.45 | CH2 | 4.4 | 2.5 | C 7, C 8, C 16, C 17 |
| C 9 | 63.2 | 61.45 | CH2 | 4.45 | 2.5 | C 7, C 8, C 16, C 17 |
| C 10 | 122.1 | 120.92 | C | |||
| C 11 | 112.1 | 111.47 | CH | 7.47 | C 10, C 12, C 13, C 16 | |
| C 12 | 148.1 | 148.25 | C | |||
| C 13 | 153.4 | 152.7 | C | |||
| C 14 | 110.4 | 110.18 | CH | 6.88 | 7.58 | C 10, C 12, C 13 |
| C 15 | 123.7 | 123.64 | CH | 7.58 | 6.88 | C 11, C 13, C 16 |
| C 16 | 166.4 | 165.72 | C | |||
| C 17 | 75.1 | 72.4 | CH | 4.45 | 2.50, 4.03 | C 7, C 8, C 9, C 16, C 18 |
| C 18 | 74.7 | 73.61 | CH2 | 4.03 | 4.45 | C 7, C 8 |
| C 19 | 101.9 | 100.9 | CH2 | 5.95 | C 3, C 4 | |
| C 20 | 56.0 | 56.3 | CH3 | 3.91 | C 12 | |
| C 21 | 56.1 | 56.3 | CH3 | 3.94 | C 13 |
The data collected in Table 1 were entered into the program and a Molecular Connectivity Diagram (MCD) was automatically created (Figure 1).
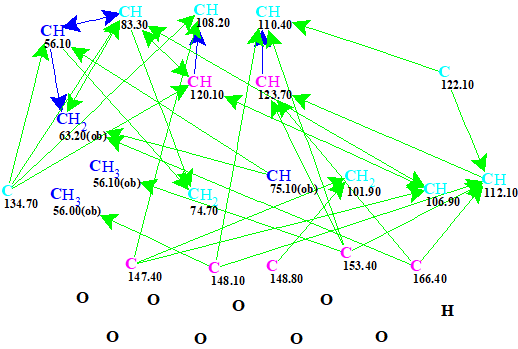
Figure 1. Molecular connectivity diagram.
No user edits of the MCD were made and it passed the contradictions checks. Therefore a Strict Structure Generation [2] accompanied with 13C chemical shift prediction was initiated.
Results: k = 72→14→11, tg = 0.3 s.
For all structures the 13C chemical shift prediction was performed using the neural networks algorithm and HOSE code based program, and then the output structural file was ranked in the order of descending dA(13C) values, the average deviations calculated by the HOSE code based program. The eight top ranked structures are presented in Figure 2.
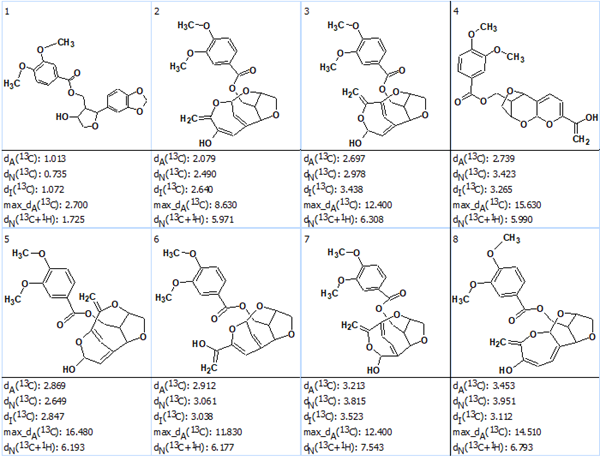
Figure 2. The eight top ranked structures of the output file.
It is seen that the structure ranked as first is identical to the structure of zanthplanispine determined in [1]. The validity of the first structure can also be seen in Figure 3 where all the carbons are colored green, which means that deviations of predicted 13C chemical shifts are less than 3 ppm.
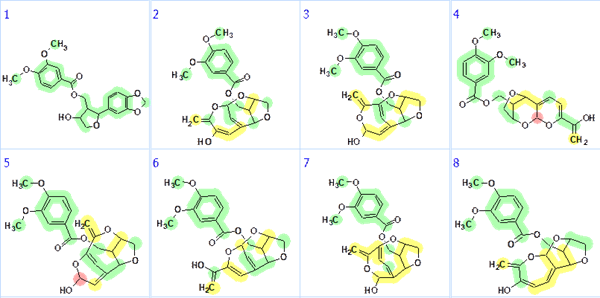
Figure 3. Top eight structures of the ranked output file. Accuracy of 13C chemical shift prediction is marked by color: green – <3 ppm, yellow – <15 ppm, red – >15 ppm.
The elucidated structure of compound 1 together with the automatically assigned 13C chemical shifts is shown below.
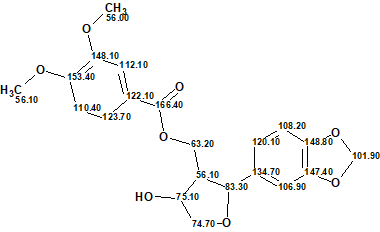
We see that the structure of this unusual tetrahydrofuran was determined by ACD/Structure Elucidator automatically without any problems in almost no time.
References
- G.-Y. Su, Y.-C. Cheng, K.-W. Wang, X.-Y. Wang, B. Wu. (2017). An Unusual Tetrahydrofuran Lignan from the Roots of Zanthoxylum planispinum and the Potential Anti-inflammatory Effects. Chem. Biodiversity, 14, e1600214.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.


