April 1, 2016
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Sarglaperoxide A
Sarcandra glabra (Thunb.) Nakai (Chloranthaceae), a folk medicinal herb widely used in China and other East and Southeast Asian countries for the treatment of inflammation and traumatic injuries, contains abundant eudesmanes, linderanes, and linderane dimers that were demonstrated to be the bioactive substances. Among them, the linderane dimers were the characteristic metabolites of the plants of Chloranthaceae, and most of them were constructed by two linderane monomers.
From investigating the constituents of Sarcandra glabra, P. Wang et al [1] isolated a pair of structurally-related terpene lactones, sarglaperoxides A and B. Structurally, these compounds present an unprecedented skeleton that is constructed by a linderane lactone and a linear normonoterpene via a six-membered ring, and features a peroxide bond between the two units. The unique structures, especially the peroxide bond, inspired authors [1] to investigate their biological activities, since many natural peroxides exhibiting significant bioactivities attributed these activities to their peroxide bonds. It is worth noting that the 2015 Nobel Prize for Physiology or Medicine was awarded to Professor Tu for her discovery of artemisinin, a famous antimalarial agent with an active peroxide bond.
The original experimental data related to Sarglaperoxide A (1) were used to challenge ACD/Structure Elucidator Suite and determine if the software could reliably elucidate this novel structure.
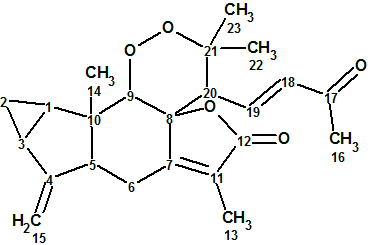
1
Sarglaperoxide A (1) was obtained as a colorless column. The molecular formula was determined as C23H28O5 by HRESIMS. NMR spectroscopic data for this compound are presented in Table 1. The 13C, 1H and HSQC spectra were transferred to Table 1 as they were tabulated in the article [1], while HMBC correlations were extracted manually from the spectrum pattern included into Supporting Information, as well as from graphical representation of key HMBC correlations in the article [1].
Table 1. Spectroscopic NMR data for Sarglaperoxide A.
| Label | δC | δC calc | XHn | δH | M | C HMBC |
| C 1 | 22.6 | 27.56 | CH | 1.91 | u | |
| C 2 | 15.9 | 15.26 | CH2 | 0.71 | u | |
| C 2 | 15.9 | 15.26 | CH2 | 0.82 | u | C 1, C 4, C 10 |
| C 3 | 23.9 | 24.22 | CH | 2.02 | u | |
| C 4 | 151 | 156.18 | C | |||
| C 5 | 53 | 54.15 | CH | 3.35 | u | |
| C 6 | 22.8 | 25.49 | CH2 | 2.35 | u | C 4, C 5, C 7, C 8, C 11 |
| C 7 | 162 | 163.57 | C | |||
| C 8 | 88.8 | 90.99 | C | s | ||
| C 9 | 88.3 | 85.15 | CH | 4.49 | u | C 5, C 8, C 10 |
| C 10 | 43.4 | 46.21 | C | |||
| C 11 | 124.9 | 126.08 | C | |||
| C 12 | 173 | 173.15 | C | |||
| C 13 | 8.6 | 8.69 | CH3 | 1.76 | s | C 7, C 11, C 12 |
| C 14 | 19.6 | 21.32 | CH3 | 0.53 | s | C 1, C 5, C 9, C 10 |
| C 15 | 107.2 | 105.72 | CH2 | 4.76 | u | |
| C 15 | 107.2 | 105.72 | CH2 | 5.07 | u | C 3, C 5 |
| C 16 | 26.6 | 26.87 | CH3 | 2.2 | s | C 17, C 18, C 19 |
| C 17 | 197.4 | 198.47 | C | |||
| C 18 | 136 | 134.82 | CH | 6.04 | d | C 20 |
| C 19 | 139.7 | 139.75 | CH | 6.35 | u | C 8, C 17 |
| C 20 | 52.2 | 49.22 | CH | 2.87 | d | C 7, C 19, C 21 |
| C 21 | 81.6 | 81.44 | C | |||
| C 22 | 21.9 | 24.34 | CH3 | 1.29 | s | C 20, C 21 |
| C 23 | 27.4 | 25.04 | CH3 | 1.42 | s | C 20, C 21 |
The data from Table 1 were input into ACD/Structure Elucidator Suite, and a Molecular Connectivity Diagram (MCD) was created by the program (see Figure 1).
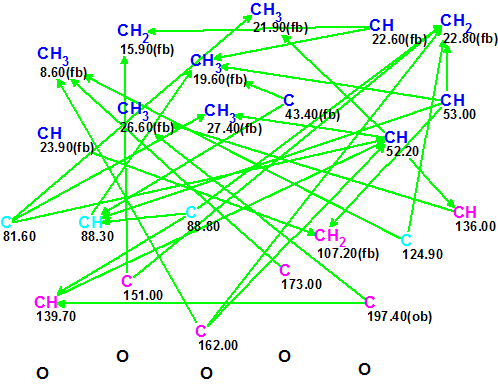
Figure 1.
Molecular Connectivity Diagram (MCD) for Sarglaperoxide A.
MCD overview. Figure 1 shows that the hybridization state of four carbon atoms (C 81.60, C 88.30, 88.80 and C 124.90) marked by light blue color was set as sp3
or sp2 (not sp). Hybridizations of sp3 and sp2 types are designated in MCD by blue and violet colors respectively. The label fb (forbidden) means that a heteroatom can not be a neighbor of a given carbon, while the label ob (obligatory) denotes the requirement of a neighboring heteroatom. Information about 1H resonances multiplicities shown in Table 1 (column M) was added to the properties of corresponding atoms.
No user edits were made to the MCD. Checking the MCD for consistency showed that there were no contradictions in 2D NMR data, and consequently Strict Structure Generation was initiated. This process was accompanied with 13C chemical shift prediction, and those structures which were characterized by average deviation d>5 ppm were automatically rejected by the system. The following results were obtained: k = 54 → 7 → 7, tg = 0.5 s. Following the general methodology [2], after 13C chemical shift prediction by three methods (Incremental, Neural Networks, HOSE code-based) the output structural file was ranked in increasing order of the average deviation dA (13C chemical shifts were calculate by HOSE code based approach). The three top structures of the ranked file are presented in Figure 2.
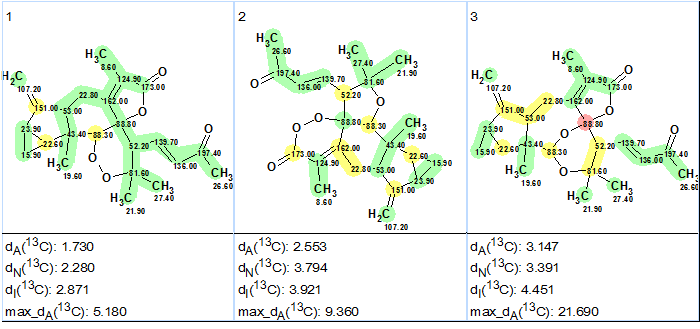
Figure 2.
The three top structures of the ranked file for Sarglaperoxide A. The color attributes of the 13C chemical shift prediction accuracy are: ±3 ppm – green, between 3 and 15 ppm – yellow, greater than 15 ppm – red.
Figure 2 shows that the top-ranked structure #1 coincides with structure of Sarglaperoxide A determined in the original article [1] and confirmed by x-ray analysis. The deviations calculated as well as the accuracy of the carbon atom chemical shift prediction marked with color allowed us to confidently conclude that the solution obtained using ACD/Structure Elucidator Suite is correct.
Finally, the elucidated structure with automatically assigned 13C chemical shifts is shown below.
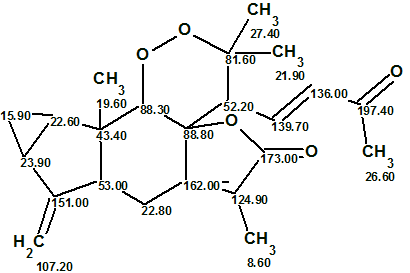
It was interesting to investigate how employing 1H multiplicities can influence the time of structure generation. The multiplicities were cancelled in the MCD, and structure generation was repeated with the same options as before, which gave the following results: k = 107 → 7, tg = 2 m 20 s. Consequently we can conclude that the usage of 1H multiplicities accelerated the structure generation process by factor of almost 300 (!) in the given example.
Thus the structure of a new natural product containing an unprecedented skeleton was robustly and instantly elucidated by the program in fully automatic mode.
References
- P. Wang, R.-J. Li, R.-H. Liu, K.-L. Jian, M.-H. Yang, L. Yang, L.-Y. Kong, J. Luo. (2016). Sarglaperoxides A and B, Sesquiterpene-Normonoterpene Conjugates with a Peroxide Bridge from the Seeds of Sarcandra glabra. Org. Lett., 18: 832–835.


