February 1, 2015
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Virosaine
Zhao and co-workers [1] isolated two new Securinega alkaloids (virosaines A and B) with an unprecedented skeleton from the twigs and leaves of Flueggea virosa and elucidated their structures. We used the spectroscopic data presented in by the authors to establish the structure of Virosaine A (structure 1) which had been confirmed by X-ray diffraction.
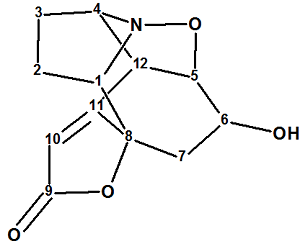
1
The molecular formula of structure 1 was established as C12H13NO4 by its HRESI-MS (m/z 258.0736 [M + Na]+, calculated for C12H13NO4Na: 258.0737). The authors proceeded with the following considerations: “The UV absorption maximum at 238 nm and IR bands at 3422, 1725, 1647 cm-1 implied the presence of an α,β-unsaturated γ-lactone ring and a hydroxyl group. The NMR spectra revealed that 1 possessed 12 carbons including an α,β-unsaturated γ-lactone ring [δH 5.84 (1H, br s); δC 175.5, 171.9, 110.8, and 85.4]. The above spectral data suggested that 1 could be a “norsecurinine-type alkaloid”. We think that such strict conclusions can be done by chemists only if they work with compounds common for a given laboratory. Instead we will try to elucidate the structure of unknown as if by an “ab initio” approach, where no information about the functional groups is known.
13C and 1H chemical shifts assigned to atoms of the molecule using HSQC were tabulated in the article, while key 1H-1H COSY and HMBC correlations were only displayed graphically on the structure. These data which were utilized for computer-assisted structure elucidation are collected in Table 1 and the Molecular Connectivity Diagram created by the program is shown in Figure 1.
Table 1: Virosaine A. Spectroscopic data
| Label | δC | δC calc | CHn | δH | M(J ) | COSY | C HMBC |
| C 1 | 74.1 | 64.6 | CH | 3.88 | u | 1.2 | C 7, C 11, C 3 |
| C 2 | 22.1 | 26.12 | CH2 | 1.2 | u | 3.88, 1.48 | |
| C 2 | 22.1 | 26.12 | CH2 | 1.7 | u | ||
| C 3 | 20.8 | 27.88 | CH2 | 1.48 | u | 1.20, 4.01 | |
| C 3 | 20.8 | 27.88 | CH2 | 1.91 | u | ||
| C 4 | 70.9 | 70.65 | CH | 4.01 | u | 3.89, 1.48 | C 5, C 11 |
| C 5 | 88 | 89.76 | CH | 4.67 | u | 4.29, 3.89 | |
| C 6 | 65.4 | 68.89 | CH | 4.29 | u | 4.67, 1.83 | C 8 |
| C 7 | 44.7 | 39.32 | CH2 | 2.92 | u | ||
| C 7 | 44.7 | 39.32 | CH2 | 1.83 | u | 4.29 | |
| C 8 | 85.4 | 90.33 | C | ||||
| C 9 | 175.5 | 172.98 | C | ||||
| C 10 | 110.8 | 111.81 | CH | 5.84 | u | ||
| C 11 | 171.9 | 172.04 | C | ||||
| C 12 | 50.2 | 47.15 | CH | 3.89 | u | 4.01, 4.67 | C 3, C 10 |
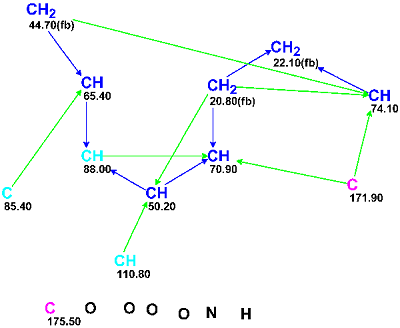
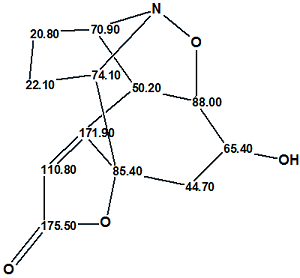
Figure 1: Molecular Connectivity Diagram for Virosaine A.
MCD overview. Four methine groups characterized by 13C chemical shifts in the range of 50.20–74.1 ppm have 1H chemical shifts falling in the chemical shift interval 3.90–4.30 ppm. Together this implies that all the mentioned carbon atoms have heteroatoms (O or N) as neighbors. Because the IR spectrum clearly indicates the presence of a carbonyl group (1725 cm-1) it would be rational to assign the carbon C 175.50 as belonging to C=O group; however the C=O bond was not drawn by hand and only the “ob” label (obligatory) was ascribed to this carbon. The chemical shift δC=171.9 ppm is also rather typical for ester or amide groups but at this stage we will leave this atom without any “ob” label (the consequences of which will be considered later). The properties of the three “light blue” carbon atoms C 85.40–C 110.80 (sp2 or sp3) were also not edited, as both carbon double bond C=C and O-C-O or O-C-N atomic groupings are possible in the molecule according to the molecular formula. The IR absorption band at 3422 cm-1 may belong to stretching vibrations both of OH and NH groups, so neither of these group could be drawn by hand on the MCD, which is shown in Figure 2 edited in accordance with these mentioned considerations.
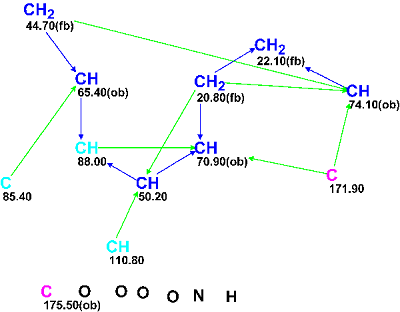
Figure 2: Edited Molecular Connectivity Diagram for Virosaine A.
MCD checking did not reveal the presence of contradictions in the 2D NMR data, and structure generation was thus run. Because chemical bonds between hereroatoms are observed relatively rare in natural products, these bonds are usually marked as not allowed in Structure Generation Options for the first run, to prevent generation of a huge number of non-realistic structures. 13C chemical shift calculations during the structure generation was activated with threshold parameters dI>4 ppm, d(max) >20 ppm, that is all structures which meet this criterion are rejected. The results were: k=2,756→0, tg= 3s; i.e., no appropriate structures were generated.
Experience showed that in such a situation the failure most frequently can be accounted for by the presence of nonstandard correlations that were not detected during the MCD checking. Therefore the next step was Fuzzy Structure Generation with initial parameters m=1, a=16, that is we introduce a suggestion that at least one NSC of unknown length exists in 2D NMR data. Results: k=75,790→0, tg = 2m 34s, 15 from 15 possible fuzzy combinations having been using during generation. The repeated failure is a hint to change the options governing the possibility of chemical bonds between heteroatoms. The next program run was initiated with an option that allowed chemical bonds between hereoatoms as shown below in Figure 3:

Figure 3: A part of a Dialog Window where chemical bonds between heteroatoms are allowed.
The results of the strict structure generation were: k=5,667→3→3, tg = 11s. The best structure selected by HOSE and incremental chemical shift predictions (dA=3.76, dI=3.13 ppm) coincided with the structure of Virosaine A (1) determined in the article. The ranked output file is shown in Figure 4 while structure 2 displays the 13C chemical shift assignments.
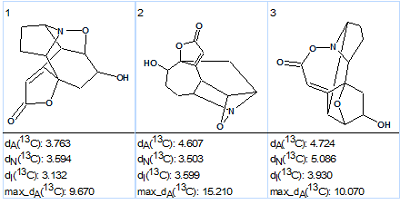
Figure. 4: The ranked output structural file for Virosaine A generated structures.
The values of deviations are relatively large, which is explained by the unprecedented structure of the molecule, but maximal deviation values also support the ranking of these 3 structures. Nevertheless the priority is not fully convincing – the difference d(#2) – d(#1) is small, and this is a good example of where CASE results should be confirmed by additional experiments (recollect that structure of Virosaine A was originally confirmed by X-ray crystallography).
Structure 2 shows that the carbon C 171.9 belonging to a carbon double bond (C)2C=C(C2) has a very unusual 13C chemical shift. It was interesting to see what will happen if this atom was supplied with an “ob” label which forces the atom to form –C=O or C=N fragments; i.e., generating structures under circumstances where structure 2 is impossible. Such a suggestion is acceptable from the point of view an unbiased expert who performs the structure elucidation “ab initio“.
2
This last run of structure generation was repeated from the MCD where carbon C 171.9 was labeled as sp2/ob. The results were: k=4,593→0, tg = 8s. By forbidding the existing bond at C 171.9, no structures are generated that are consistent with the experimental data, and this negative result confirms that structure 2 is the correct one, even with the unusual chemical shift at this position.
References
- Zhao BX, Wang Y, Zhang DM, Huang XJ, Bai LL, Yan Y, Chen JM, Lu TB, Wang YT, Zhang QW, Ye WC. (2012). Virosaines A and B, two new birdcage-shaped Securinega alkaloids with an unprecedented skeleton from Flueggea virosa. Org Lett. 14(12):3096–3099.


