March 1, 2013
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Indol alkaloid
Dried roots and leaves of the Isatis indigotica plant are used in traditional Chinese medicine for the treatment of various diseases. Diverse structures and significant biological activities from extracts of this plant have attracted considerable interest. Chemical and pharmacological studies have resulted in the characterization of constituents with different structural features and biological activities. As part of a program to assess the chemical and biological diversity of traditional Chinese medicines, an aqueous extract of the roots of I. indigotica has been investigated by Chen and co-workers.1 In this work, the authors isolated and structurally characterized an indole alkaloid containing unusual dihydrothiopyran and 1,2,4-thiadiazole rings (1).
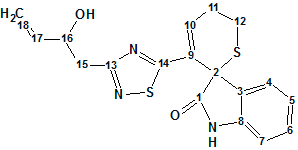
1
Herein, we will discuss structure elucidation of compound 1 using the ACD/Structure Elucidator software. The IR spectrum of 1 showed absorption bands for hydroxy and/or amino groups (3256 cm-1), carbonyl (1715 cm-1), and aromatic ring (1619 and 1474 cm-1) functionalities. The positive mode ESIMS of 1 exhibited quasimolecular ion peaks at m/z 372 [M+H]+, 394 [M+Na]+, and 410 [M+K]+. The molecular formula of C18H17N3O2S2, with 12 degrees of unsaturation, was determined from HRESIMS at m/z 372.0844 [M + H]+ (calculated for C18H17N3O2S2, 372.0835) and 394.0659 [M+Na]+ (calculated for C18H17N3O2S2Na, 394.0654), combined with the NMR data which are presented in Table 1.
Table 1. Indole alkaloid. Spectroscopic NMR data.
| Label | δC | δC calc | XHn | δH | M(1H) | COSY | C HMBC |
| C 1 | 177.1 | 174.5 | C | ||||
| C 2 | 49.5 | 55.32 | C | ||||
| C 3 | 130.3 | 132.07 | C | ||||
| C 4 | 125.2 | 125.35 | CH | 7.07 | u | 6.9 | C 2, C 6, C 8 |
| C 5 | 122.9 | 121.64 | CH | 6.9 | u | 7.24, 7.07 | C 7, C 3 |
| C 6 | 130.5 | 128.42 | CH | 7.24 | u | 6.90, 6.99 | C 8, C 4 |
| C 7 | 110.9 | 109.05 | CH | 6.99 | u | 7.24 | C 3, C 5 |
| C 8 | 143.7 | 142.13 | C | ||||
| C 9 | 128.6 | 129.41 | C | ||||
| C 10 | 142.1 | 134.7 | CH | 7.4 | dd(4.8, 4.2) | 2.84 | C 2, C 12, C 14 |
| C 11 | 27.8 | 26.07 | CH2 | 2.84 | u | 7.40, 3.76 | C 9 |
| C 12 | 22.2 | 23.86 | CH2 | 2.74 | u | ||
| C 12 | 22.2 | 23.86 | CH2 | 3.76 | u | 2.84 | C 2, C 10 |
| C 13 | 174.1 | 165.95 | C | ||||
| C 14 | 187.3 | 183.41 | C | ||||
| C 15 | 41.5 | 39.11 | CH2 | 2.88 | u | 4.31 | C 17, C 13 |
| C 15 | 41.5 | 39.11 | CH2 | 2.79 | u | ||
| C 16 | 71.1 | 71.65 | CH | 4.31 | u | 5.63, 2.88, 3.95 | C 18, C 13 |
| C 17 | 141.5 | 139.27 | CH | 5.63 | u | 4.98, 4.31 | C 15 |
| C 18 | 114 | 115.4 | CH2 | 4.98 | u | 5.63 | C 16 |
| N 1 | 100* | NH | 9.68 | u | C 1, C 2, C 3, C 8 | ||
| O 1 | 150* | OH | 3.95 | u | 4.31 | C 15, C 17 |
* Fictitious 15N and 17O NMR chemical shifts.
Molecular Connectivity Diagram (MCD) overview. Three carbon atoms—C 110.9, C 128.6 and C 130.3—marked by the program as “sp2 or sp3” were defined as “sp2” hybridized. The methine group CH 71.1(4.31), as well as quaternary carbons C 174.1–C 187.3, were labeled as “ob” for obligatory. Two hydrogen atoms were allowed to be attached to neighbors of carbon C 142.1 (see Table 1). The MCD was then checked by the software, which noted that at least one non-standard connectivity exists in the collective 2D NMR data. It is worthwhile to note that the existence of chemical bonds between heteroatoms was not suggested during the MCD checking.
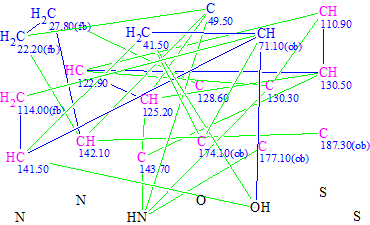
Figure 1. Indole alkaloid. Slightly edited Molecular Connectivity Diagram.
Structures were then generated by the software using Fuzzy Structure Generation, with “Choose Option Automatically” selected, followed by 13C chemical shift calculations The following results were obtained: k=163→1, tg=2 s , one connectivity was extended (m=1); one single suitable structure was returned in two seconds. The single structure 2 characterized with deviations dA=5.650 , dI=3.132 , dN=3.583 , dA(max)=30.210 ppm is shown below:
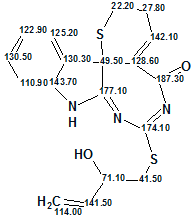
2
Deviations of value 3.1–3.6 ppm are not uncommon when the analyzed structure is unusual, but the value dA=5.65 ppm, together with dA(max)= 30.21 ppm (associated with the chemical shift 177.1 ppm) can be considered as a warning, hinting at the necessity of additional computational experiments. Therefore the Fuzzy Structure Generation was repeated with the options m=2, a=1. Results: k=1731→2, tg=52 s, with 465 from 465 possible connectivity combinations being checked. From this run, structure 2 was again generated, along with a second structure for which deviations turned out significantly larger.
Because that the deviations do not decrease with higher m values, we decided to check the possibility of forming chemical bonds between heteroatoms (until now the software assumed that these bonds are forbidden). With this in mind, the “Check MCDs Options” were set as shown in Figure 2.
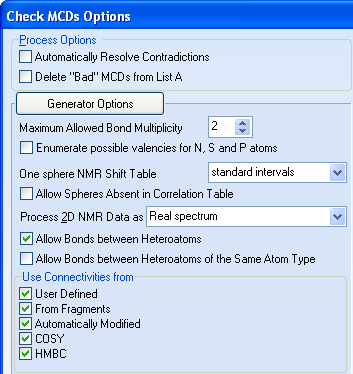
Figure 2. Indole alkaloid Options of the structure generation. Checkbox “Allow Bonds between Heteroatoms” is marked.
No contradictions were found in MCD now, and strict structure generation was performed with the following results: k=780→313→313, tg=0.4 s. The three top ranked structures of the output file are presented in Figure 3.
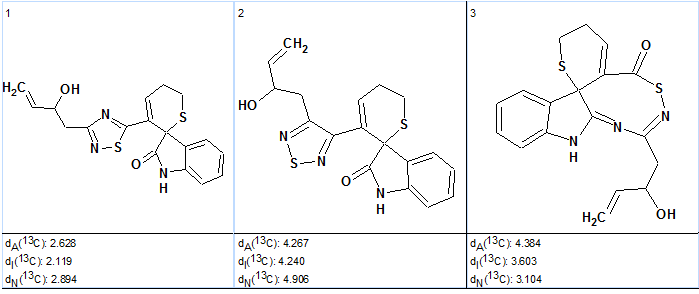
Figure 3. Indole alkaloid. Three top ranked structures of the output file.
The best structure (#1, left) is found to coincide with the structure 1, and the 13C chemical shift assignments are displayed on structure 1a:

1a
This example demonstrates that using the normal problem solving strategies of ACD/Structure Elucidator, a structure possessing fairly unusual skeleton can be quickly and reliably (D=d(2) – d(1) @ 2 ppm) identified.
References
- M. Chen, S. Lin, L. Li, C. Zhu, X. Wang, Y. Wang, B. Jiang, S. Wang, Y. Li, J. Jiang, J. Shi. Enantiomers of an Indole Alkaloid Containing Unusual Dihydrothiopyran and 1,2,4-Thiadiazole Rings from the Root of Isatis indigotica. Org. Lett., 14(22): 5668–5671, 2012.


