June 1, 2017
by Mikhail Elyashberg, Leading Researcher, ACD/Labs
Spiroschincarin A
Schisandra incarnate is a form of a woody liana. It is mainly found in the west and southwest of Hubei province, China. Its fruit has found many uses in traditional Chinese medicine like treating contusion and rheumatism. Song et al [1] investigated the chemical constituents of the fruit of S. incarnata. They isolated five novel spirocyclic compounds, spiroschincarins A−E, featuring a unique 1-oxaspiro[6.6]tridecane moiety. Their structures, with absolute configurations, were determined by extensive spectroscopic studies, single-crystal X-ray diffraction, and experimental ECD. The structure elucidation of spiroschincarin A (1) from NMR data was used for challenging ACD/Structure Elucidator.
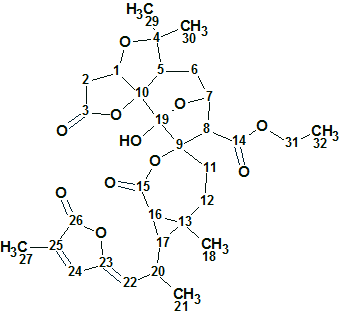
1
Spiroschincarin A (1) was isolated as colorless needles. The molecular formula, C31H38O11, was determined by HRESIMS (m/z 609.2334, [M + Na] +, calcd. for 609.2312) and 13C NMR data. The molecule has 13 degrees of unsaturation. The IR spectrum showed the presence of a hydroxyl group (3428 cm−1) and carbonyl groups (1714 and 1748 cm−1).
In the article [1], 13C and 1H NMR data were presented as tables while key HMBC and COSY correlations were presented in a graphic form (Figure 1). There was no complete table with the HMBC and COSY correlations.
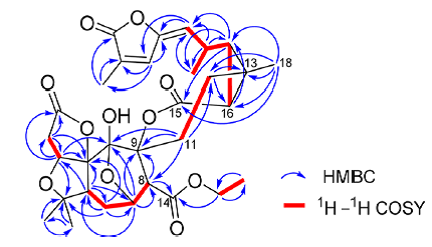
Figure 1. Spiroschincarin A. Key HMBC and COSY correlations.
The established molecular formula as well as the data from the tables and from Figure 1 were entered into the program (Table 1).
Table 1. Spiroschincarin A. Spectroscopic NMR data.
| Label | δC | δCcalc* | CHn | δH | M | COSY | H to C HMBC |
| C 1 | 80.9 | 75.63 | CH | 5.04 | u | 2.49 | C 2, C 3, C 10, C 19 |
| C 2 | 40.3 | 36.97 | CH2 | 2.49 | u | 5.04 | C 1, C 3, C 10 |
| C 2 | 40.3 | 36.97 | CH2 | 2.92 | u | ||
| C 3 | 179.4 | 173.3 | C | ||||
| C 4 | 86.7 | 86.58 | C | ||||
| C 5 | 47.4 | 42.61 | CH | 3.11 | u | 2.67 | C 6, C 7, C 10 |
| C 6 | 29.7 | 30.06 | CH2 | 1.95 | u | ||
| C 6 | 29.7 | 30.06 | CH2 | 2.67 | u | 3.11, 4.45 |
C 4, C 5, C 7, C 10 |
| C 7 | 74.9 | 70.37 | CH | 4.45 | u | 2.67, 3.31 |
C 5, C 8, C 9, C 19 |
| C 8 | 57.5 | 59.5 | CH | 3.31 | d | 4.45 | C 9, C 14 |
| C 9 | 93.2 | 93.1 | C | ||||
| C 10 | 99.5 | 96.91 | C | ||||
| C 11 | 36.5 | 33.42 | CH2 | 2.45 | u | ||
| C 11 | 36.5 | 33.42 | CH2 | 2.42 | u | 1.89 | C 8, C 9, C 12, C 19 |
| C 12 | 24.6 | 31.52 | CH2 | 1.52 | u | ||
| C 12 | 24.6 | 31.52 | CH2 | 1.89 | u | 2.42 | C 13, C 16 |
| C 13 | 28.7 | 29.97 | C | ||||
| C 14 | 168.9 | 168.2 | C | ||||
| C 15 | 173.8 | 172.88 | C | ||||
| C 16 | 32.4 | 27.58 | CH | 1.77 | u | 1.33 | C 17 |
| C 17 | 40.6 | 34.91 | CH | 1.33 | u | 1.77, 2.65 |
C 4, C 21, C 22, C 29 |
| C 18 | 26.3 | 20.41 | CH3 | 1.12 | s | C 12, C 13, C 15, C 16, C 17 | |
| C 19 | 105.2 | 110.11 | C | ||||
| C 20 | 31 | 31.09 | CH | 2.65 | u | 1.22, 1.33, 5.53 |
C 17, C 21, C 22 |
| C 21 | 20.2 | 21.71 | CH3 | 1.22 | d | 2.65 | C 22 |
| C 22 | 119.2 | 117.97 | CH | 5.53 | d | 2.65 | C 23, C 24 |
| C 23 | 148.7 | 146.65 | C | ||||
| C 24 | 137.3 | 138.65 | CH | 7.58 | s | C 23, C 25, C 27 | |
| C 25 | 130.9 | 128.84 | C | ||||
| C 26 | 172.8 | 171.15 | C | ||||
| C 27 | 10.6 | 10.76 | CH3 | 1.92 | u | C 26 | |
| C 29 | 25.4 | 21.63 | CH3 | 1.27 | s | C 4, C 30 | |
| C 30 | 30.2 | 28.11 | CH3 | 1.33 | s | ||
| C 31 | 61.8 | 60.75 | CH2 | 4.25 | u | 1.3 | C 14, C 32 |
| C 31 | 61.8 | 60.75 | CH2 | 3.99 | u | ||
| C 32 | 14.4 | 14.08 | CH3 | 1.3 | t | 4.25 | C 31 |
* 13C chemical shift prediction was carried out using HOSE code based approach [2].
A slightly edited Molecular Connectivity Diagram (MCD) is presented in Figure 2. Inspection of Figure 1 shows that there is an HMBC correlation from the hydrogens of methyl group C 18 to carbon C15, which is of 4 bonds length. Examining the HMBC spectrum presented in the Supporting Information of [1] we see that this peak is very weak. For this reason we manually define this correlation to be of 2-4 bonds in length (pink arrow in Figure 2).
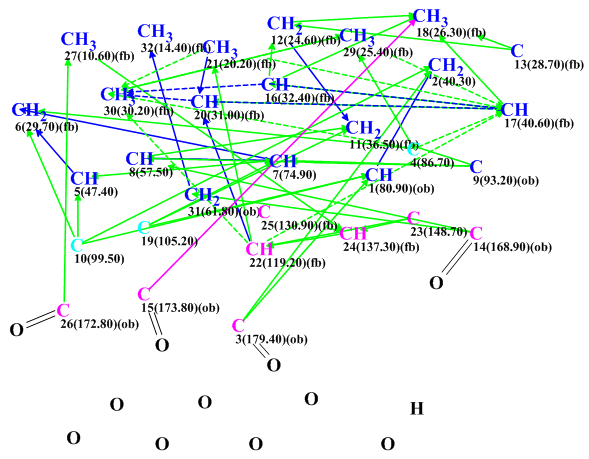
Figure 2. Spiroschincarin A. Molecular Connectivity Diagram.
MCD overview. Since only key HMBC correlations presented in Figure 1 were entered into the program, it was appropriate to introduce some additional information to MCD to help accelerate the structure generation. In this respect the four evident carbonyl groups were manually defined and the two carbons, C 99.5 and C 105.2 (colored in blue), were set to be of an ambiguous hybridization “not sp” (sp2 or sp3) as formation of double bonds C=C and O-C-O groups is possible in the presence of a large number of oxygen atoms. Multiplicities of all CH3 groups were set in according to data presented in Table 1. All the above edits were very evident from the spectra and would not have caused any unintentional bias in the structure generation. Strict structure generation accompanied by 13C chemical shift prediction (threshold averaged deviation dI = 5 ppm) was initiated and was completed with the following results: k=13,272 → 1064 →1064, tg = 4 m 38 s.
As is common for ACD/Structure Elucidator 13C NMR chemical shift prediction, three approaches (increments, neural networks and HOSE code based methods) were followed, and the output file was ranked in increasing order of dA values (average deviations of HOSE codes calculated chemical shift from experimental ones). The six top ranked structures are shown in Figure 3.
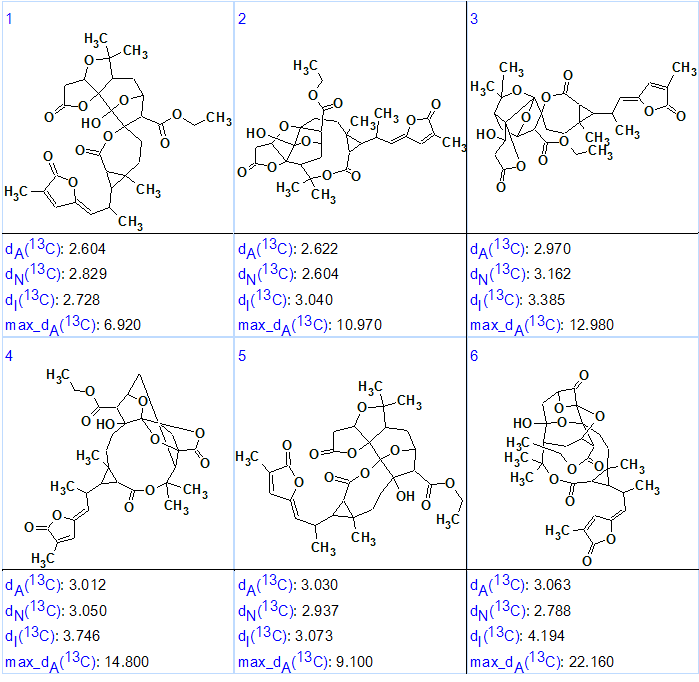
Figure 3. Spiroschincarin A. The top six ranked structures of the output file.
We see that structure #1 is characterized by the smallest average deviations calculated by HOSE codes and incremental methods of NMR spectrum prediction [2]. Comparison of this structure with the published formula of Spiroschincarin A shows that both structures are identical. However since the difference between the dA values for the first two ranked structures (#1 and #2) is small, the validity of the first structure would need additional confirmation by other methods. Structure #1 was confirmed, though, by X-ray analysis in [1]. In addition, structure #2 looks somewhat unique, with an 11-atom lactone ring. Further proof could have probably been obtained if full HMBC and COSY data were available.
The elucidated structure with automatically assigned 13C chemical shifts is shown below.
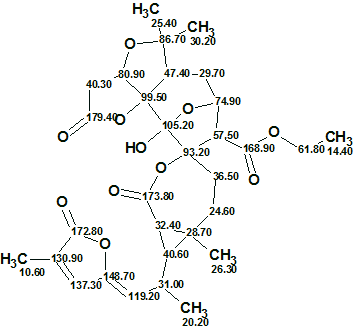
In conclusion the structure of a new complex molecule containing a unique fragment was elucidated within less than 5 minutes with incomplete HMBC and COSY data and found to be in agreement with the published structure which was also confirmed by X-ray diffraction.
A complete elucidation package that speeds up the elucidation process and ensures that no candidate is overlooked. Learn more about ACD/Structure Elucidator Suite.
References:
- J. Song, Y. Liu, M. Zhou, H. Cao, X.-G. Peng, J.-J. Liang, X.-Y. Zhao, M. Xiang, H.-L. Ruan. (2017). Spiroschincarins A−E: Five Spirocyclic Nortriterpenoids from the Fruit of Schisandra incarnata. Org. Lett. 19: 1196−1199.
- M.E. Elyashberg, A.J. Williams. (2015). Computer-based Structure Elucidation from Spectral Data (p. 454). Springer-Verlag Berlin, Heidelberg.
For more hands-on experience in the methods of Computer-Assisted Structure Elucidation, using ACD/Structure Elucidator, we recommend the textbook Computer–Based Structure Elucidation from Spectral Data from Springer, by M.E. Elyashberg and A.J. Williams.
The special version of ACD/Structure Elucidator and 100 problems (in electronic format) which were discussed in the book are available at www.acdlabs.com/TeachingSE.


